The mDOT Center
Transforming health and wellness via temporally-precise mHealth interventions






mDOT@MD2K.org
901.678.1526
901.678.1526








Collaborating Investigator:
Dr. Daniel E. Rivera (PI), Arizona State University
Funding Status:
NIH/NLM
3/6/19 – 2/28/23
Associated with:
IEEE/ACM international conference on Connected Health: Applications, Systems and Engineering Technologies (CHASE)
September 12, 2022
Bayesian inference, probabilistic programming, time series, missing data, Bayesian imputation, mobile health
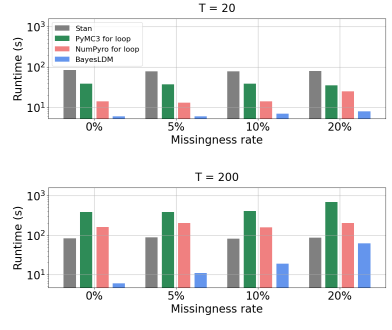
We have developed a toolbox for the specification and estimation of mechanistic models in the dynamic bayesian network family. This toolbox focuses on making it easier to specify probabilistic dynamical models for time series data and to perform Bayesian inference and imputation in the specified model given incomplete data as input. The toolbox is referred to as BayesLDM. We have been working with members of CP3, CP4, and TR&D2 to develop offline data analysis and simulation models using this toolbox. We are also currently in discussions with members of CP4 to deploy the toolbox’s Bayesian imputation methods within a live controller optimization trial in the context of an adaptive walking intervention.
In this paper we present BayesLDM, a system for Bayesian longitudinal data modeling consisting of a high-level modeling language with specific features for modeling complex multivariate time series data coupled with a compiler that can produce optimized probabilistic program code for performing inference in the specified model. BayesLDM supports modeling of Bayesian network models with a specific focus on the efficient, declarative specification of dynamic Bayesian Networks (DBNs). The BayesLDM compiler combines a model specification with inspection of available data and outputs code for performing Bayesian inference for unknown model parameters while simultaneously handling missing data. These capabilities have the potential to significantly accelerate iterative modeling workflows in domains that involve the analysis of complex longitudinal data by abstracting away the process of producing computationally efficient probabilistic inference code. We describe the BayesLDM system components, evaluate the efficiency of representation and inference optimizations and provide an illustrative example of the application of the system to analyzing heterogeneous and partially observed mobile health data.
We present a a toolbox for the specification and estimation of mechanistic models in the dynamic bayesian network family.
Journal of Biomedical Informatics, Vol. 158
October 2024
System identification, Idiographic modeling, Dynamical systems modeling, Physical activity, Behavior change, Wearables
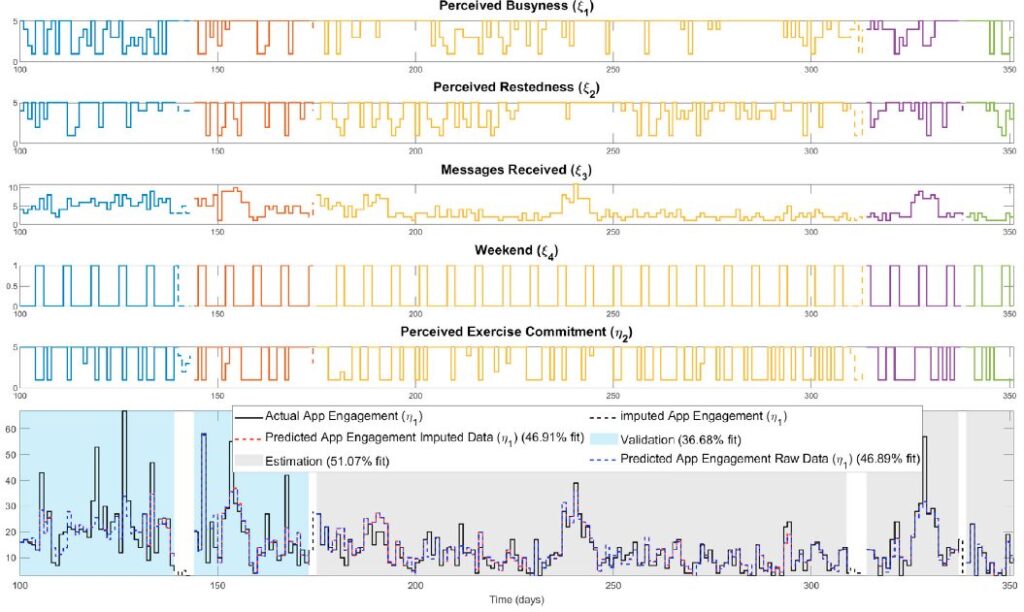
Digital behavior change interventions (DBCIs) are feasibly effective tools for addressing physical activity. However, in-depth understanding of participants’ long-term engagement with DBCIs remains sparse. Since the effectiveness of DBCIs to impact behavior change depends, in part, upon participant engagement, there is a need to better understand engagement as a dynamic process in response to an individual’s ever-changing biological, psychological, social, and environmental context. The year-long micro-randomized trial (MRT) HeartSteps II provides an unprecedented opportunity to investigate DBCI engagement among ethnically diverse participants. We combined data streams from wearable sensors (Fitbit Versa, i.e., walking behavior), the HeartSteps II app (i.e. page views), and ecological momentary assessments (EMAs, i.e. perceived intrinsic and extrinsic motivation) to build the idiographic models. A system identification approach and a fluid analogy model were used to conduct autoregressive with exogenous input (ARX) analyses that tested hypothesized relationships between these variables inspired by Self-Determination Theory (SDT) with DBCI engagement through time.
Data from 11 HeartSteps II participants was used to test aspects of the hypothesized SDT dynamic model. Across individuals, the number of daily notification prompts received by the participant was positively associated with increased app page views. The weekend/weekday indicator and perceived daily busyness were also found to be key predictors of the number of daily application page views. This novel approach has significant implications for both personalized and adaptive DBCIs by identifying factors that foster or undermine engagement in an individual’s respective context. Once identified, these factors can be tailored to promote engagement and support sustained behavior change over time.
This research explored long-term engagement with Digital Behavior Change Interventions (DBCIs) for physical activity using data from the year-long HeartSteps II trial. By combining data from wearables, app usage, and motivation assessments, the study developed models to understand engagement as a dynamic process. Key findings showed that daily notification prompts, whether it was a weekend or weekday, and perceived daily busyness were significant predictors of how often participants viewed the app pages. This novel approach has important implications for creating personalized and adaptive DBCIs that can better foster and sustain user engagement over time.
IFAC-PapersOnLine, Vol. 58, Iss. 15
October 2024
eHealth, Bayesian methods, Computational Social Sciences, Time series modelling
Digital behavior change interventions (DBCIs) such as “just-in-time” adaptive interventions (JITAIs) have demonstrated efficacy for increasing physical activity behavior. However, the effectiveness of these interventions is heavily dependent upon user engagement. Despite the inherent dynamic nature of engagement, as it varies over time based on an individual’s changing environment, context, and psychological state, the current understanding of engagement primarily comes from static snapshots of the behavior. The availability of intensive longitudinal data from JITAIs provides a unique opportunity to build and test dynamic models of behavior change from a process systems lens, relying on prediction-error methods from system identification. However, data missingness is a significant practical consideration in this process. Therefore, in this work we address missingness using a Bayesian imputation approach, which we evaluate using data from the HeartSteps II JITAI. Ultimately, the methods presented support the discovery of key factors that impact engagement behavior over time and can play an important role in the development of large-scale personalized interventions.
This publication focuses on using system identification and Bayesian imputation to create dynamic models of user engagement in mHealth digital behavior change interventions (DBCIs), such as “just-in-time” adaptive interventions (JITAIs). The goal is to move beyond static views of engagement to understand how it changes over time, addressing the practical issue of data missingness with a Bayesian approach. This research aims to identify key factors influencing engagement and support the development of personalized interventions, demonstrated using data from the HeartSteps II JITAI.
Martín Moreno
Advisor
Daniel E. Rivera
Committee Members
Arizona State University
August 2016
mHealth, behavioral interventions, Social Cognitive Theory (SCT), system identification, control engineering, physical inactivity, Hybrid Model Predictive Control (HMPC), Identification Test Monitoring (ITM), adaptive interventions, cyberphysical systems.
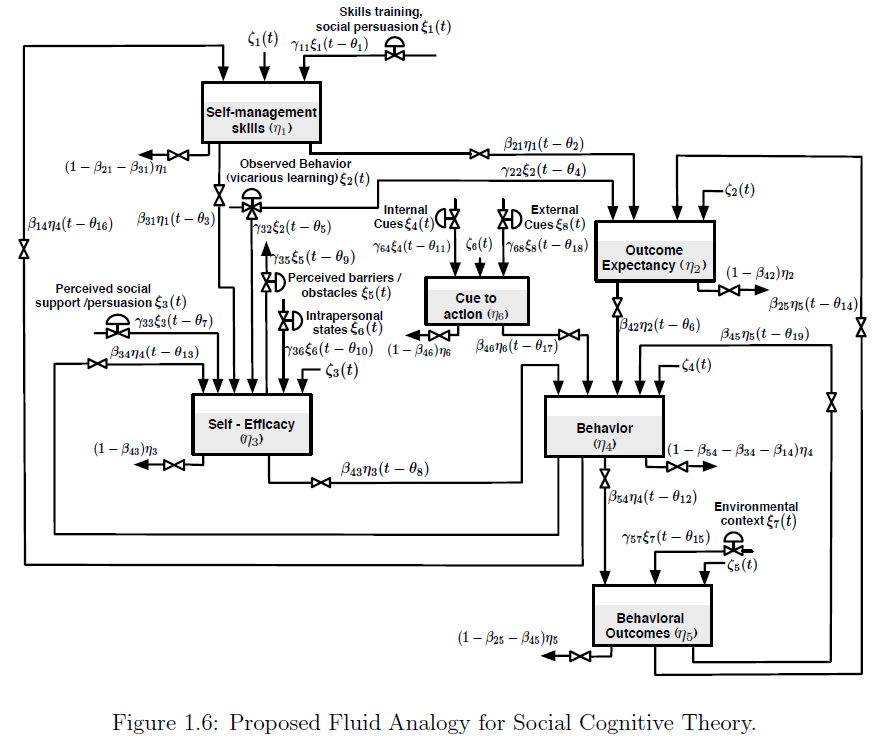
Behavioral health problems such as physical inactivity are among the main causes of mortality around the world. Mobile and wireless health (mHealth) interventions offer the opportunity for applying control engineering concepts in behavioral change settings. Social Cognitive Theory (SCT) is among the most influential theories of health behavior and has been used as the conceptual basis of many behavioral interventions. This dissertation examines adaptive behavioral interventions for physical inactivity problems based on SCT using system identification and control engineering principles. First, a dynamical model of SCT using fluid analogies is developed. The model is used throughout the dissertation to evaluate system identification approaches and to develop control strategies based on Hybrid Model Predictive Control (HMPC). An initial system identification informative experiment is designed to obtain basic insights about the system. Based on the informative experimental results, a second optimized experiment is developed as the solution of a formal constrained optimization problem. The concept of Identification Test Monitoring (ITM) is developed for determining experimental duration and adjustments to the input signals in real time. ITM relies on deterministic signals, such as multisines, and uncertainty regions resulting from frequency domain transfer function estimation that is performed during experimental execution. ITM is motivated by practical considerations in behavioral interventions; however, a generalized approach is presented for broad-based multivariable application settings such as process control. Stopping criteria for the experimental test utilizing ITM are developed using both open-loop and robust control considerations.
A closed-loop intensively adaptive intervention for physical activity is proposed relying on a controller formulation based on HMPC. The discrete and logical features of HMPC naturally address the categorical nature of the intervention components that include behavioral goals and reward points. The intervention incorporates online controller reconfiguration to manage the transition between the behavioral initiation and maintenance training stages. Simulation results are presented to illustrate the performance of the system using a model for a hypothetical participant under realistic conditions that include uncertainty. The contributions of this dissertation can ultimately impact novel applications of cyberphysical system in medical applications.
This dissertation presents a system identification and control engineering approach to optimize mobile health (mHealth) behavioral interventions, specifically addressing physical inactivity. It develops a dynamical model of Social Cognitive Theory (SCT) using fluid analogies, which is then used to evaluate system identification methods and develop control strategies based on Hybrid Model Predictive Control (HMPC). The work also introduces Identification Test Monitoring (ITM) procedures to determine the shortest necessary experimental duration while ensuring sufficient data for accurate model identification. Ultimately, these contributions aim to impact novel applications of cyberphysical systems in medical contexts.
CP4 will initially focus on the analysis of the HeartSteps v1 data set using a Model-on-Demand estimation procedure for inferring just-in-time states. The primary proximal outcomes in HeartSteps v1 is 30-minute step count, as recorded by a Jawbone fitness tracker. This device has the property that if the device is worn and no steps are detected, no step count is recorded. Thus, both a zero-step count and missing step count (e.g., due to lack of connectivity, device not being worn, device not being on, etc.) appear as missing data in the HeartSteps v1 data set. Initial treatment efficacy analysis performed using the HeartSteps v1 data set was based on zero imputation of missing step values, discarding the uncertainty due to missingness. The methods developed under TR&D1 Aim 1 will be applied to the HeartSteps v1 data set to model and represent the uncertainty due to missing step count information. While CP3 will test the clinical utility of uncertainty model in interventions in its MRT, CP4 will assess the utility of the TR&D1 in dynamical systems modeling over multiply imputed data sets. The two teams will work together to improve the missing step count models and analyze the impact of missing step count modeling on end-to-end inference of just-in-time states. The powerful modeling framework developed in CP4 will provide a rigorous and well-specified setting in which to create and refine uncertainty models through multiple push-pull iterations between the two teams.
You must be logged in to post a comment.
No Comments