The mDOT Center
Transforming health and wellness via temporally-precise mHealth interventions






mDOT@MD2K.org
901.678.1526
901.678.1526








Collaborating Investigator:
Dr. Zach Ives, University of Pennsylvania (PI: Dr. Santosh Kumar, University of Memphis)
Funding Status:
ACI-1640183
NSF/ACI
9/01/16 – 8/31/21
Associated with:
Konstantinos Mamouras, Mukund Raghothaman, Rajeev Alur, Zachary G. Ives, Sanjeev Khanna
Proceedings of the 38th ACM SIGPLAN Conference on Programming Language Design and Implementation
December 2017
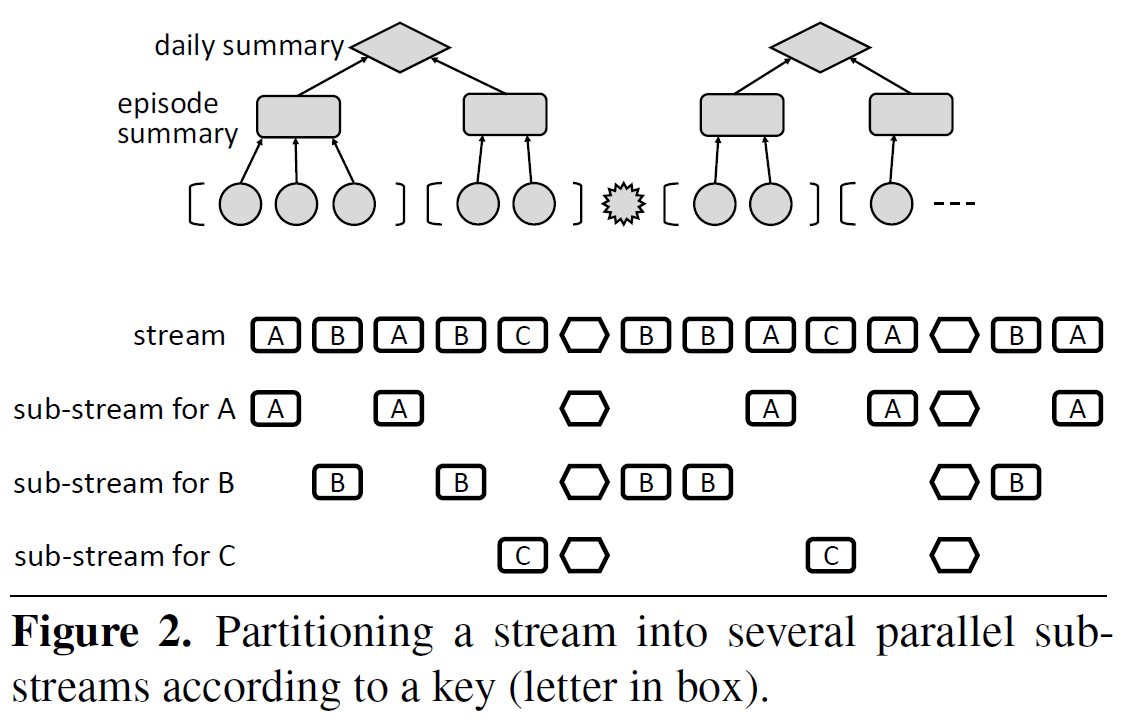
Data stream processing, Quantitative Regular
Expressions, IoT applications, real-time decision making, relational query languages, regular expressions, modular specification, efficient
evaluation, streaming composition, key-based
partitioning, pattern-based windows, approximate aggregation.
Real-time decision making in emerging IoT applications typ-ically relies on computing quantitative summaries of largedata streams in an efficient and incremental manner. To sim-plify the task of programming the desired logic, we proposeStreamQRE, which provides natural and high-level constructsfor processing streaming data. Our language has a novel in-tegration of linguistic constructs from two distinct program-ming paradigms: streaming extensions of relational querylanguages and quantitative extensions of regular expressions.The former allows the programmer to employ relational con-structs to partition the input data by keys and to integrate datastreams from different sources, while the latter can be used toexploit the logical hierarchy in the input stream for modularspecifications.We first present the core language with a small set ofcombinators, formal semantics, and a decidable type system.We then show how to express a number of common patternswith illustrative examples. Our compilation algorithm trans-lates the high-level query into a streaming algorithm withprecise complexity bounds on per-item processing time andtotal memory footprint. We also show how to integrate ap-proximation algorithms into our framework. We report onan implementation in Java, and evaluate it with respect toexisting high-performance engines for processing streamingdata. Our experimental evaluation shows that (1) StreamQREallows more natural and succinct specification of queriescompared to existing frameworks, (2) the throughput of ourimplementation is higher than comparable systems (for ex-ample, two-to-four times greater than RxJava), and (3) theapproximation algorithms supported by our implementationcan lead to substantial memory savings.
StreamQRE is a novel programming language designed for real-time decision-making in IoT applications by integrating two powerful paradigms: streaming extensions of relational query languages and quantitative extensions of regular expressions. This allows for natural and modular specification of complex queries over large data streams. Its compiler translates high-level queries into efficient streaming algorithms with guaranteed low memory footprint and fast processing times, outperforming existing high-performance streaming engines like RxJava, Esper, and Flink. StreamQRE also supports approximation algorithms for significant memory savings on computationally intensive tasks like median calculation.
Bo-Jhang Ho, Nima Nikzad, Bharathan Balaji, Mani Srivastava
Proceedings of the 2017 ACM International Joint Conference on Pervasive and Ubiquitous Computing
Proceedings of the 2017 ACM International Symposium on Wearable Computers
December 2017
Context-aware, mobile
applications, engagement,
just-in-time assessment, push notifications, user studies, mhealth, framework
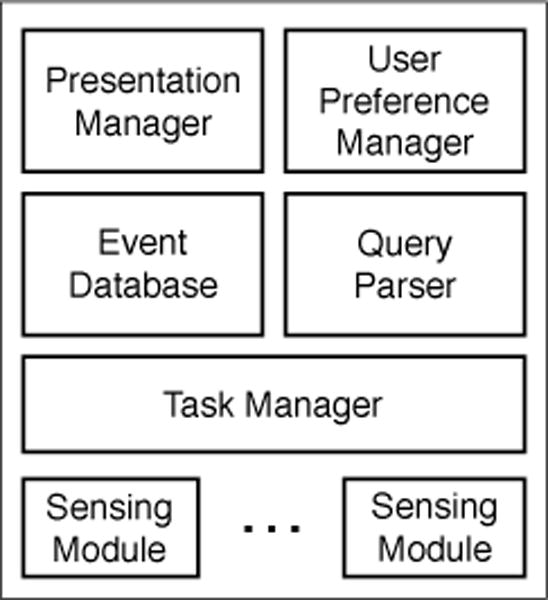
Mobile technologies that drive just-in-time ecological momentary assessments and interventions provide an un-precedented view into user behaviors and opportunities to manage chronic conditions. The success of these meth-ods rely on engaging the user at the appropriate moment, so as to maximize questionnaire and task completion rates. However, mobile operating systems provide little support to precisely specify the contextual conditions in which to notify and engage the user, and study designers often lack the ex-pertise to build context-aware software themselves. To ad-dress this problem, we have developed Emu, a framework that eases the development of context-aware study appli-cations by providing a concise and powerful interface for specifying temporal- and contextual-constraints for task no-tifications. In this paper we present the design of the Emu API and demonstrate its use in capturing a range of scenar-ios common to smartphone-based study applications.
Emu is a framework designed to simplify the development of context-aware mobile study applications. It provides a concise and powerful interface for specifying temporal and contextual constraints for task notifications, addressing the challenge that mobile operating systems offer limited support for precisely engaging users at appropriate moments. By automating the tracking of contextual states and user responses, Emu maximizes questionnaire and task completion rates in studies, helping to manage chronic conditions and improve study protocol adherence. It significantly reduces code complexity for developers compared to native implementations.

Proceedings of the 2017 Workshop on Wearable Systems and Applications
June 19, 2017
Mobile sensors can track human states, the surrounding context, daily behaviors, and exposures to environmental risk factors in the natural field environment. Real-time analysis of such sensor data makes it possible to deliver personalized recommendations to improve health, wellness, and performance. Widely used GPS-navigation systems that provide just-in-time directions for traffic-aware navigation and activity trackers that help users set and achieve daily physical activity goals are widely used early examples. The increasing availability of mobile sensors that allow collection of raw sensor data, along with mobile big data software platforms that allow labeled collection, curation, modeling, and visualization of such data for development and validation of new markers and sensor-triggered interventions, is opening up exciting new research directions. They include novel sensor systems for selftracking of health, wellness, and performance.
Proceedings of the 2024 CHI Conference on Human Factors in Computing Systems (CHI ’24)
May 11, 2024
stress-tracking, stressor-logging, visualizations, behavioral changes, stress intervention, emotion/affective computing, wearable sensors, field studies, empirical studies in HCI, longitudinal study, self-reflection, personal informatics, stress reduction, self-awareness, smartwatch apps, smartphone apps, physiological events, and daily stressors.
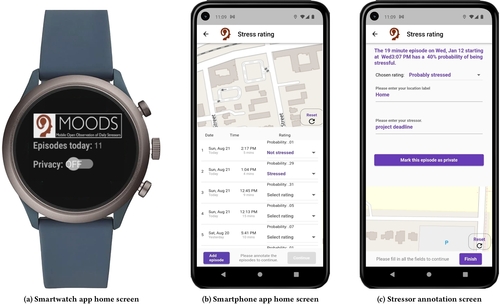
Commercial wearables from Fitbit, Garmin, and Whoop have recently introduced real-time notifications based on detecting changes in physiological responses indicating potential stress. In this paper, we investigate how these new capabilities can be leveraged to improve stress management. We developed a smartwatch app, a smartphone app, and a cloud service, and conducted a 100-day field study with 122 participants who received prompts triggered by physiological responses several times a day. They were asked whether they were stressed, and if so, to log the most likely stressor. Each week, participants received new visualizations of their data to self-reflect on patterns and trends. Participants reported better awareness of their stressors, and self-initiating fourteen kinds of behavioral changes to reduce stress in their daily lives. Repeated self-reports over 14 weeks showed reductions in both stress intensity (in 26,521 momentary ratings) and stress frequency (in 1,057 weekly surveys).
A 100-day field study (MOODS) explored how wearables can enhance stress management by combining real-time physiological stress detection with momentary stressor logging and weekly self-reflective visualizations. The study found significant reductions in self-reported stress intensity and frequency, fostering greater self-awareness of stressors and prompting participants to make 14 types of self-initiated behavioral changes for improved emotional regulation, productivity, and self-care. This work highlights the potential of personal informatics systems in driving lasting stress reduction through user engagement and data insights.
Syed Monowar Hossain, Timothy Hnat, Nazir Saleheen, Nusrat Jahan Nasrin, Joseph Noor, Bo-Jhang Ho, Tyson Condie, Mani Srivastava, Santosh Kumar
Proceedings of the 15th ACM Conference on Embedded Network Sensor Systems
November 6, 2017
mHealth, mobile sensor big data, software architecture, digital biomarkers, wearable sensors
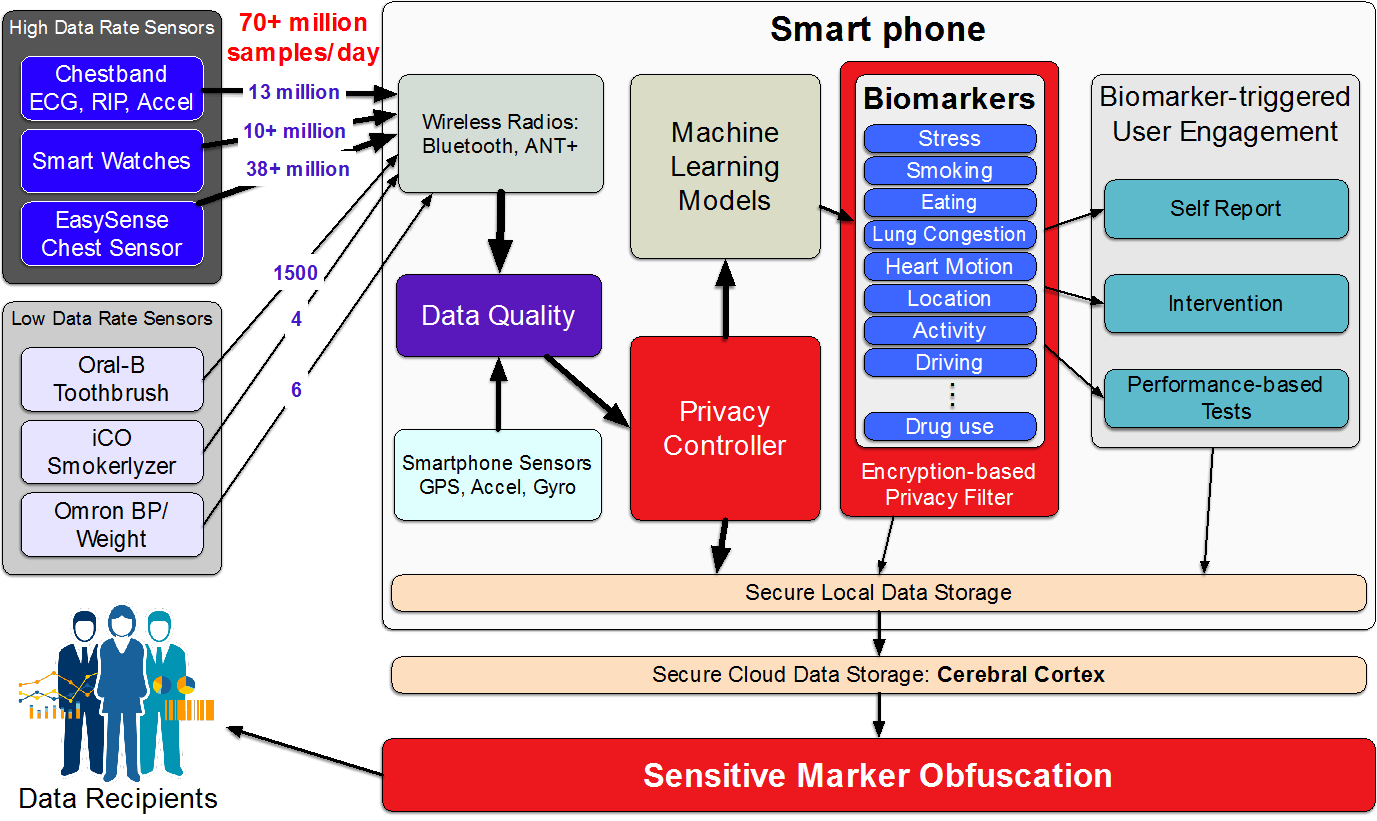
The development and validation studies of new multisensory biomarkers and sensor-triggered interventions requires collecting raw sensor data with associated labels in the natural field environment. Unlike platforms for traditional mHealth apps, a software platform for such studies needs to not only support high-rate data ingestion, but also share raw high-rate sensor data with researchers, while supporting high-rate sense-analyze-act functionality in real-time. We present mCerebrum, a realization of such a platform, which supports high-rate data collections from multiple sensors with realtime assessment of data quality. A scalable storage architecture (with near optimal performance) ensures quick response despite rapidly growing data volume. Micro-batching and efficient sharing of data among multiple source and sink apps allows reuse of computations to enable real-time computation of multiple biomarkers …
mCerebrum is an open-source mobile sensing software platform designed for the development and validation of digital biomarkers and sensor-triggered interventions. Unlike traditional mHealth apps, it’s built to handle high-rate raw sensor data collection in natural environments, supporting over 70 million samples per day. The platform features a scalable storage architecture (Pebbles) with near-optimal performance, an efficient data exchange architecture (DataKit) for real-time biomarker computation, and a reconfigurable, burden- and context-aware scheduler for participant prompts. With a modular design spanning over 23 apps, mCerebrum has evolved through its use in scientific field studies at ten sites, accumulating 106,806 person-days of data. Evaluations show it significantly outperforms other platforms like AWARE, HealthKit, and Google Fit in data rates, storage throughput, and CPU usage, enabling efficient processing of multi-sensor biomarkers.
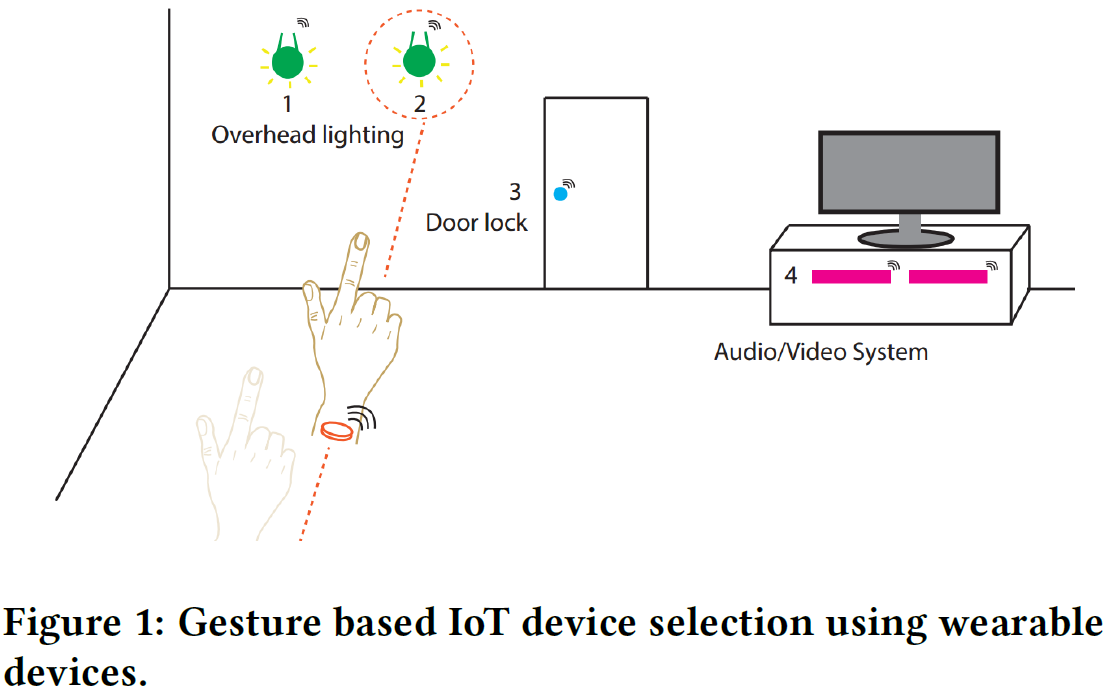
Amr Alanwar, Moustafa Alzantot, Bo-Jhang Ho, Paul Martin, Mani Srivastava
Proceedings of the Second International Conference on Internet-of-Things Design and Implementation
April 2017
Pointing, IoT, hand gestures, smartwatch, UWB, device selection, smart home, human-computer interaction (HCI), gesture recognition, wearable devices.
Although different interaction modalities have been proposed in the field of human-computer interface (HCI), only a few of these techniques could reach the end users because of scalability and usability issues. Given the popularity and the growing number of IoT devices, selecting one out of many devices becomes a hurdle in a typical smarthome environment. Therefore, an easy-to-learn, scalable, and non-intrusive interaction modality has to be explored. In this paper, we propose a pointing approach to interact with devices, as pointing is arguably a natural way for device selection. We introduce SeleCon for device selection and control which uses an ultra-wideband (UWB) equipped smartwatch. To interact with a device in our system, people can point to the device to select it then draw a hand gesture in the air to specify a control action. To this end, SeleCon employs inertial sensors for pointing gesture detection and a UWB transceiver for identifying the selected device from ranging measurements. Furthermore, SeleCon supports an alphabet of gestures that can be used for controlling the selected devices. We performed our experiment in a 9m-by-10m lab space with eight deployed devices. The results demonstrate that SeleCon can achieve 84.5% accuracy for device selection and 97% accuracy for hand gesture recognition. We also show that SeleCon is power efficient to sustain daily use by turning off the UWB transceiver, when a user’s wrist is stationary.
SeleCon is a novel system that enables natural and scalable interaction with Internet of Things (IoT) devices in smart homes using pointing gestures and hand gestures. Users wear a custom smartwatch equipped with Ultra-Wideband (UWB) and inertial sensors. By pointing to a device, the system identifies the target using UWB ranging data, and then hand gestures drawn in the air control the selected device. SeleCon prioritizes energy efficiency by only activating the power-hungry UWB when wrist motion indicates a potential pointing action. The system demonstrates high accuracy: 84.5% for device selection and 97% for hand gesture recognition.
There are major hurdles to using mobile sensor data to advance research on computational modeling of human health and behavior, including lack of access to high-quality mobile sensor data, regulatory obligations in accessing and using mobile sensor data collected from humans, and a lack of metadata capture and access services for the provenance, quality, and integrity of the data and inferences made from it. CP7 is developing a new cyberinfrastructure called mProv to annotate high-frequency mobile sensor data with data source, quality, validity, and semantics to facilitate the sharing of such data with the wider research community for third party research. It is developing techniques to integrate metadata and data capture over mobile streaming data, and propagate such data in order to enable reasoning about uncertainty and variability; runtime infrastructure and APIs for efficient sensor data acquisition and reply (integrated with human data capture), and mechanisms for managing privacy policies. To support interpretation of sensor-derived features and inferences (i.e., markers of health, behavior, and context) by researchers (for concurrent development that makes use of datastreams developed by other researchers) and automating analysis by machines, CP7 has developed datastream representation to support a common metadata structure that allows both mCerebrum and Cerebral Cortex (installed on mobile phones and the cloud respectively) to annotate the datastream with metadata. It has also developed storage, interface, instrumentation, and visualization tools for provenance tracking through stream processing operators. Provenance information can be automatically captured as a series of entities, activities, and relationships in a graph database from which it can be queried or visualized, even in near-real-time. It has built a core provenance repository with user authentication, group creation, and metadata storage capabilities. These are exposed through a simple REST microservices framework, and they can be retargeted at the back-end to a variety of SQL and NoSQL database systems. Currently, CP7 software uses Cassandra, REDIS, Neo4J, and Postgres. CP7 also works closely with an R24 from NIBIB (R24EB025845; PI: Ida Sim, UC San Francisco; 7/1/17-6/30/20) to standardize biomarkers that have been validated and being adopted in the research community via an IEEE Working Group (P1752, Open Mobile Health Standards). To evaluate its work under realistic settings, CP7 is conducting multiple iterations of 100-day field studies in 100 participants in collaboration with the Open Humans project to generate open data set that can be used by researchers to develop mHealth biomarkers for detecting daily stressors.
CP7 will get access to novel biomarkers from TR&D3 Aim 1 so that it can develop and implement appropriate annotations, provenance, and pursue standardization. This step will enable the adoption of these biomarkers by the wider research community, especially among researchers working on secondary analysis of existing datasets. A particularly relevant category of metadata that CP7 is concerned with is that relating to privacy. These include metadata describing context-dependent privacy policy that govern downstream sharing and use of sensor measurements and derivative micromarkers and biomarkers. It also includes sharing of metadata capturing transformation (e.g. sanitization, addition of noise etc.) that data stream may have undergone due to upstream exercise of privacy policy so as to allow robust computation of biomarkers that discriminate between privacy related data quality degradation and data missingness or degradation due causes such as battery exhaustion, network connectivity outages, sensor detachments, and others. As the data collected in CP7 studies will be publicly available and capture large amounts of sensor data in daily life, CP7 offers a tremendous opportunity for bi-directional interaction with TR&D3’s research under Aim 3. On the one hand, the privacy mechanisms developed under TR&D3 Aim 3 will provide CP7 with concrete instances of privacy related transformation that the metadata framework must capture. On the other hand, the metadata framework in CP7 and its stressors studies provides a vehicle for the TR&D3 Aim 3’s methods and tools for privacy-utility tradeoff in biomarker computation to be evaluated at scale and made available to the community in the form of concrete implementation. The interaction with CP7 regarding metadata mechanisms will be an iterative push-pull, whereby the feedback from deployment of the mechanisms in the studies will be used to refine the privacy mechanisms under Aim 3, which in turn will be used by CP7 to refine the metadata framework and re-evaluated in a new iteration of the Open Humans study.
You must be logged in to post a comment.
No Comments